DG Tutorial
Rather than starting from scratch, this tutorial is designed to introduce anyone interested to the problem of Domain Generalization and assist whoever wants to get into the field navigate the literature.
Introduction
Domain Generalization is a fundamental problem in machine learning today. Despite the fact that deep learning models have seen immense success in the past few years even surpassing experts in some cases they often fail to mimic the adaptability prowess of humans. The development of highly generalizable and robust ML models proves to be exceptionally difficult in numerous cases, as the distribution shifts present across separate datasets or databases cause a model’s performance to deteriorate, or even completely break down, when evaluated on previously unseen data. Most ML models depend on the assumption that the test samples are independent from and identically distributed (i.i.d.) with the training data. In practice, even though the independence assumption commonly holds, the test data often follows a different data generating distribution than the training data. This creates a need for the development of algorithms which are not affected by the distributional shift present in distinct data domains of the same problem - or in other words, domain generalizable models.
This short tutorial serves as an introduction to the field of Domain Generalization and presents its most important aspects, from problem statement, to state-of-the-art methods and evaluation settings.
Presenters
- Christos Diou, Assistant Professor of Artificial Intelligence and Machine Learning, Harokopio University of Athens, cdiou@hua.gr
- Aristotelis Ballas, PhD Student, Harokopio University of Athens, aballas@hua.gr
Outline
- Introduction to DG
- Overview of representative DG methods
- DG datasets and benchmarks
- DG for biomedical signals: The BioDG benchmark
- Conclusions
- Appendix
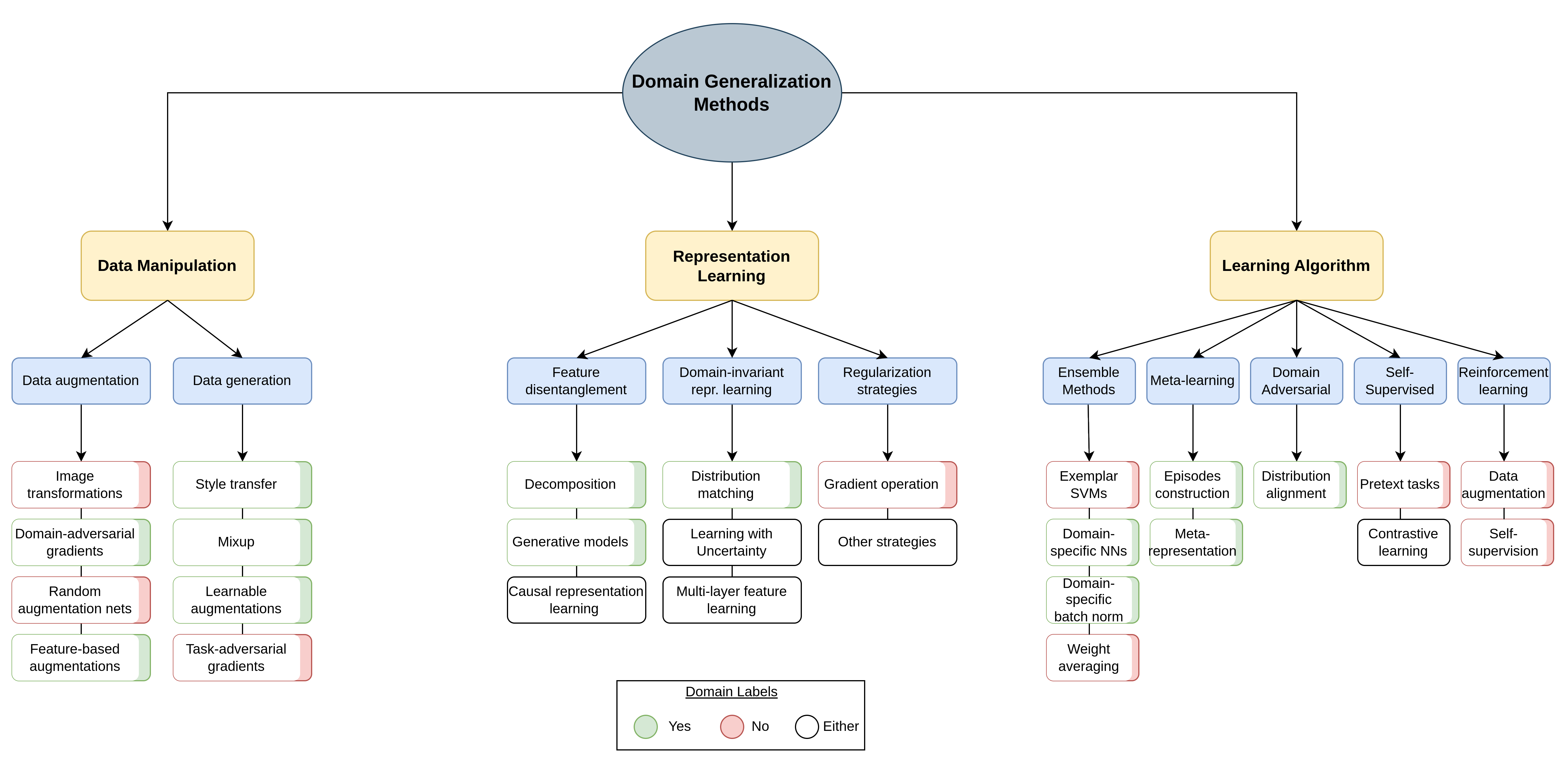
Below you may find a taxonomy of the domain generalization methods proposed in the literature.
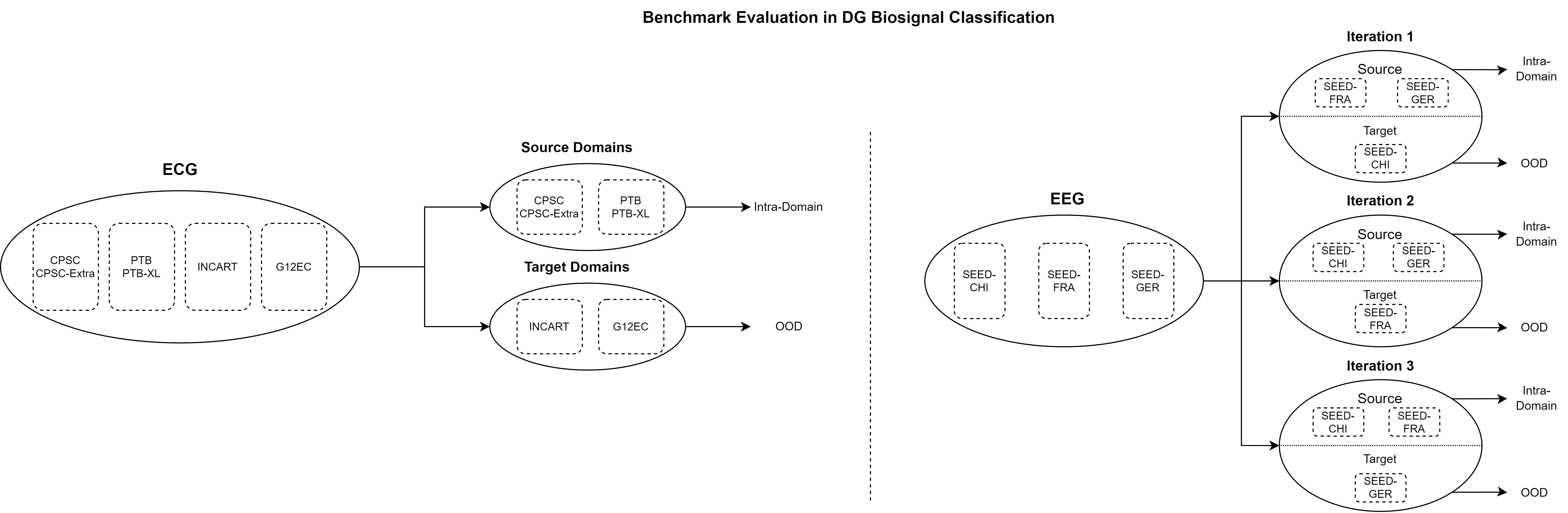
BioDG: A Benchmark for Domain Generalization in Biosignal Classification
We introduce an open-source DG evaluation benchmark, namely BioDG, for 12-lead ECG and 62-channel EEG biosignals. BioDG adapts SoTA DG algorithms from the image classification domain for 1D biosignal classification.
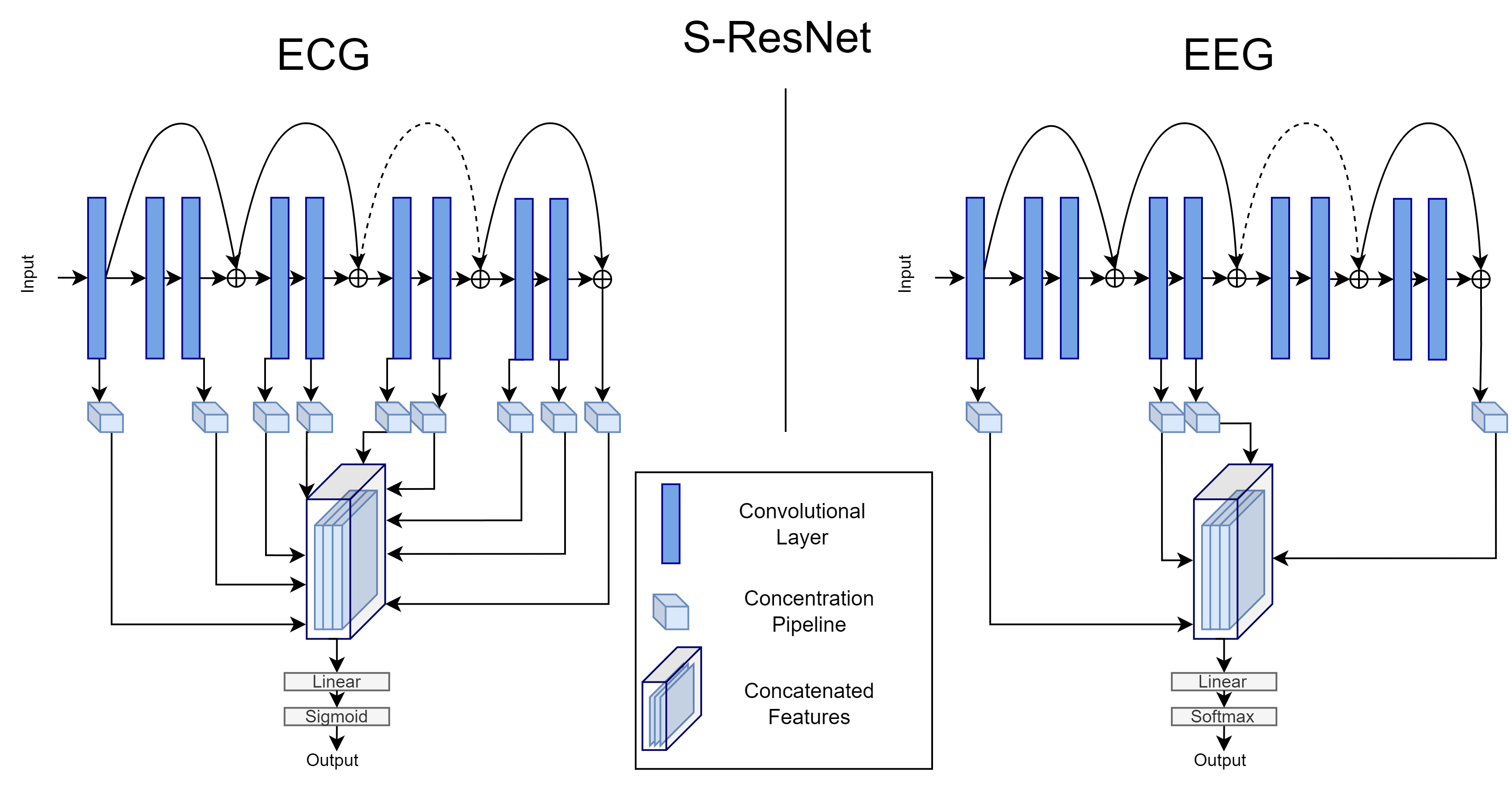
We propose an alternative neural network architecture which leverages intermediate representations from multiple layers of a CNN.
If you want to find out more about our work, please visit the following links:
Acknowledgment
The work leading to these results has been funded by the European Union under Grant Agreements No. 101057821, project RELEVIUM and European Union’s Horizon 2020 research and innovation programme under Grant Agreement No. 965231, project REBECCA (REsearch on BrEast Cancer induced chronic conditions supported by Causal Analysis of multisource data).


